Worth knowing about. For its Medicare Advantage plans, United Healthcare needs to follow Medicare policies, and for genomics, that's mostly local or LCD policies.
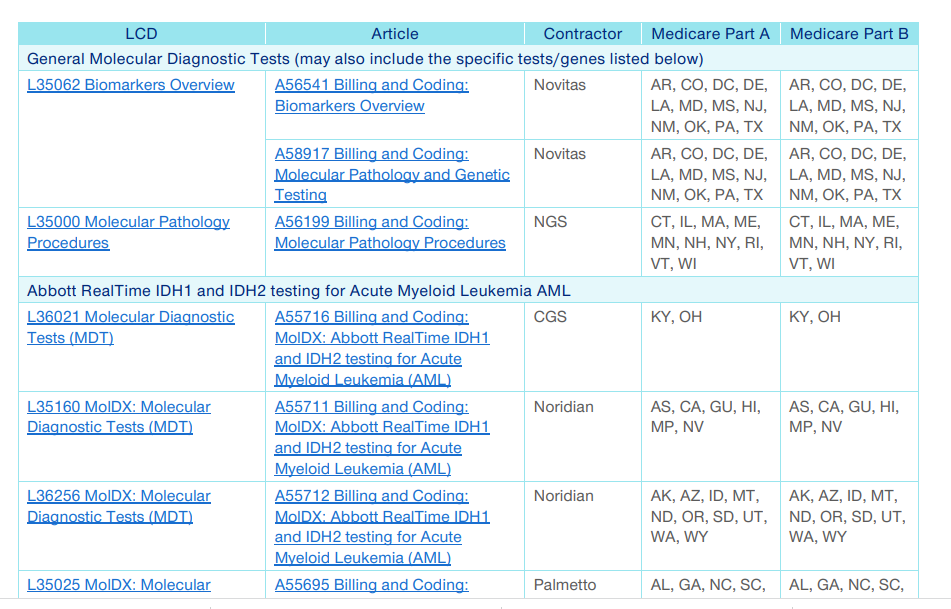
United Healthcare publishes a 49 page guide, updated regularly, of which pages 5-49 are an index to the numerous local LCDs and billing articles. Handy to know about.
MolDx Defines an Algorithm
An algorithm being defined as....
_______
An algorithm may be considered a meaningful and independent component of a laboratory process when ALL the following conditions are met:
- It is an unambiguous problem-solving operation that includes deploying a set of rules or calculations requiring computer processing;
- The test result (or a component of the result) is the calculated output of this process, and not an intermediary process;
- The same or similar test result could not be obtained without the use of this process;
- The input for the computation is derived from biological samples using analytical processes, and must include data from the sample submitted for the test;
- The process must:
- Either be required for the analytical result, OR
- If adjunct to the analytical result as a post-analytical process, the calculation itself must be independently found to be reasonable and necessary apart from the other components of the test.
Examples:
- A gene expression profile test wherein sequencing data must be compared in a calculation to an existing and validated set of profiles to bin it in one of several possible risk stratification groups would require the use of an algorithm as defined above.
- A next generation sequencing (NGS) test that uses computation to identify variants in a sample is not considered as using an algorithm in this context. The calculation in this scenario is seen as an intermediary process.
Calculations using only clinical information not derived from analytical services on biological samples are not considered algorithms in this context.
Examples would include using the clinical information from the patient in a calculation to assess their risk stratification or using a similar process to identify relevant clinical annotations derived from literature as associations with sequencing variants.
A test that inputs resultant analytical processes that are reasonable and necessary (such as gene variants or protein markers) that are post processed by computation, but wherein that subsequent computation is not independently established as reasonable and necessary above and beyond the other lab components, shall not be considered an algorithm as a valid component of a laboratory test.